Science with Passion
Application No.: VPH0078 Version 1 / 03/2024
High-throughput and sensitive HPLC analysis of lipids used in LNP formulations with evaporative light scattering detection
M. Prüfer, G. Greco; pruefer@knauer.net
KNAUER Wissenschaftliche Geräte GmbH, Hegauer Weg 38, 14163 Berlin, www.knauer.net
Illustration: curapath.com
Summary
Lipid nanoparticles (LNPs) are used to administer mRNA vaccines against Covid-19 and are considered as a crucial factor in ending the pandemic. Moreover, these nanoparticles are being researched as a potential delivery system for active pharmaceutical ingredients (API) that can combat infectious diseases, cancer, and genetic disorders. KNAUER is the global leader in the production of impingement jet mixing systems for the formulation of LNPs and also provides HPLC systems which can be used for the analysis and quality control of LNP formulations. Here, we show the development of several analytical methods for high-throughput analysis of the lipid composition of LNPs. The lipids can be analyzed with a fully porous or a core-shell phase in gradient or isocratic mode. The separation was speed up to a 2 min method. KNAUER AZURA® UHPLC system coupled to evaporative light scattering detection provided high sensitivity, with LODs in the range of 0.8–3 μg/ml (4–15 ng) and LOQs of 0.9–4 μg/ml (5–32 ng).
Introduction
Lipid nanoparticles (LNPs) became famous for the rapid development and large-scale production of vaccines during the Covid-19 pandemic. These vaccines consist of mRNA coding for an antigen encapsulated in LNPs. KNAUER helped to combat the pandemic by delivering impingement jet mixing systems enabling the formulation of LNPs in research scale, for pre-clinical and clinical trials, as well as for the large-scale manufacturing of vaccines (for more information visit (https://www.knauer.net/lipid-nanoparticle-production). More recently, LNPs gained interest as delivery systems for RNA as a therapeutic drug beyond vaccines, e.g. for cancer treatment or the treatment of inherited disorders [1]. In research and in pharmaceutical production, LNPs are subjected to careful quality control. The identity, quality, and purity of the raw materials, especially the mRNA and lipids, as well as the quality and composition of the final product must be verified. Many of the control criteria can be assayed using high performance liquid chromatography (HPLC).
In this application note we show the development of HPLC methods developed with KNAUER AZURA® HPLC system for assessing two quality criteria of LNPs: the identity of lipids and the lipid content [2]. As the lipids used in LNP manufacturing lack a suitable chromophore for detection by UV adsorption, the versatile evaporative light scattering detector (ELSD) is used for this method. The ELSD can detect most analytes which are less volatile than the used eluent. As the phenyl-hexyl column appeared to be the best choice as a stationary phase in literature [3, 4], two columns were tested here: one with a core-shell material and one with a fully porous material. For method development, a quaternary low pressure gradient pump was used. All method parameters were chosen under consideration of easy method transfer to a binary LC-MS system.
Sample Preparation
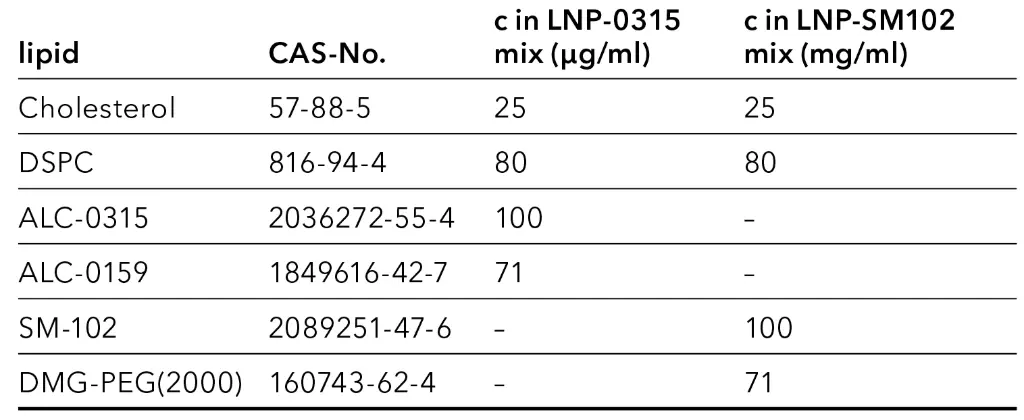
The lipids used in two LNP formulations for mRNA vaccines were provided as lipid nanoparticle exploration kits LNP-0315 and LNP-102 by Cayman Chemical. All analytes were dissolved in gradient grade ethanol or provided as a solution. The solutions were handled only with glass syringes and stored in glass vials. LNP-0315 mix contained the lipids of the vaccine BNT162b2 developed by Biontech-Pfizer, which are Cholesterol, the phospholipid 1,2-Distearoyl-sn-glycero-3-PC (DSPC), the PEGylated lipid ALC-0159 and the ionizable lipid ALC-0315. LNP-SM102 mix consisted of the lipids of the vaccine mRNA-1273 from Moderna. Besides Cholesterol and DSPC, it contained the ionizable lipid SM-102 and PEGylated lipid DMG-PEG(2000). Mixtures were prepared in ethanol:water 90:10 v/v. Dilutions and calibration standards were produced by diluting the mixtures in ethanol:water 90:10 v/v. The composition of the stock solutions of both lipid mix standards are shown in Tab. 1.
Tab. 1 Composition of lipid mix stock solutions
Results
Method development with a core-shell phenyl-hexyl stationary phase
According to the literature, various columns, eluents, modifiers and gradients can be applied to separate the lipids present in LNP formulations. For method development, a short column with core-shell particles and a phenyl-hexyl stationary phase was chosen. The column temperature (CT) was set to 50°C and ELSD evaporation temperature (ET) to 45°C. Some simple approaches for the gradient were tested first. Initially, a water-acetonitrile gradient with ammonium acetate as modifier was tested to separate the lipids of the Biontech-Pfizer vaccine. A short gradient separating all four lipids was found, but the peakshape was not satisfactory (Fig. 1). Especially ALC-0159 eluted as a very broad peak, whereas DSPC and ALC-0315 showed strong tailing.
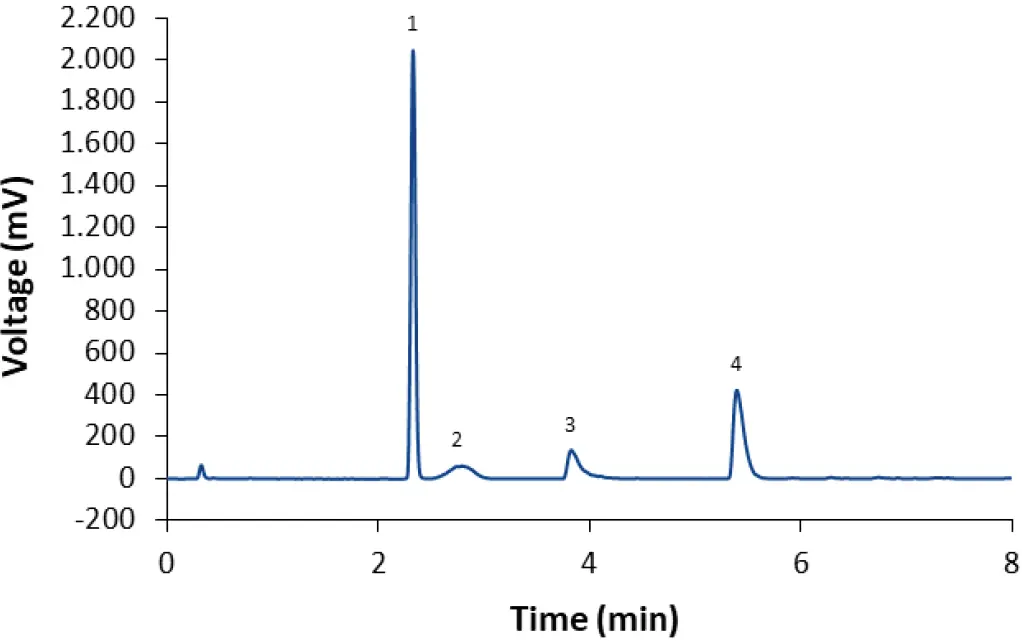
Fig. 1 Separation of LNP-0315 mix using core-shell phenyl-hexyl column and a gradient with acetonitrile (70 % 90 % in 4 min). CT = 50°C; ET = 45°C; 0.4 ml/min; 10 mM ammonium acetate as modifier. 1: Cholesterol; 2: ALC-0159; 3: DSPC; 4: ALC-0315.
By using methanol instead of acetonitrile as eluent, the peak shape for DSPC and ALC-0315 was improved, although both analytes elute later than in the acetonitrile gradient (Fig. 2). Unfortunately, the PEGylated lipid ALC-0159 does not elute as a detectable peak, but as a very flat ramp at the end of the isocratic hold (6–8 min).
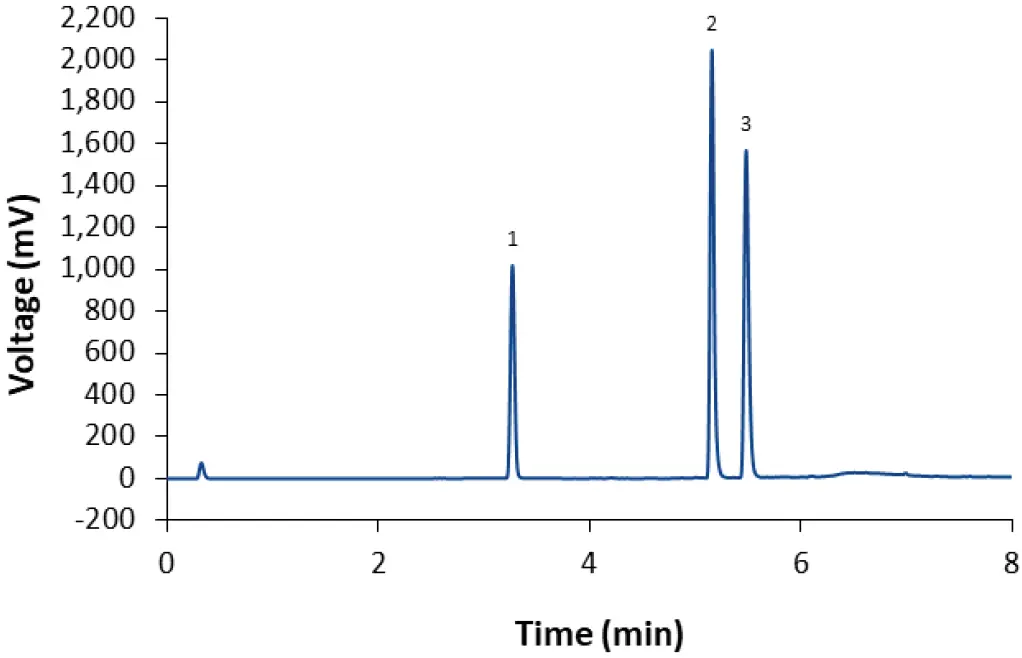
Fig. 2 Separation of LNP-0315 mix using core-shell phenyl-hexyl column and a gradient with methanol (80 % 95 % in 4 min). CT = 50°C; ET = 45°C; 0.4 ml/min; 10 mM ammonium acetate as modifier. 1: Cholesterol; 2: DSPC; 3: ALC-0315.
By combining the organic solvents, both issues can be resolved. This can be achieved with a gradient which gradually replaces methanol by acetonitrile, keeping a constant water and modifier content [5]. A similar method was used to separate the lipids of the LNP-encapsuled RNA drug Patisiran and can be transferred to a binary pump by premixing each organic solvent with water and the modifier ammonium acetate [5]. After adapting the column temperature, a very similar method was used to separate both lipid mixtures examined here (Fig. 3, Fig. 4). All lipids are eluted in 4 min and show a good peak shape.
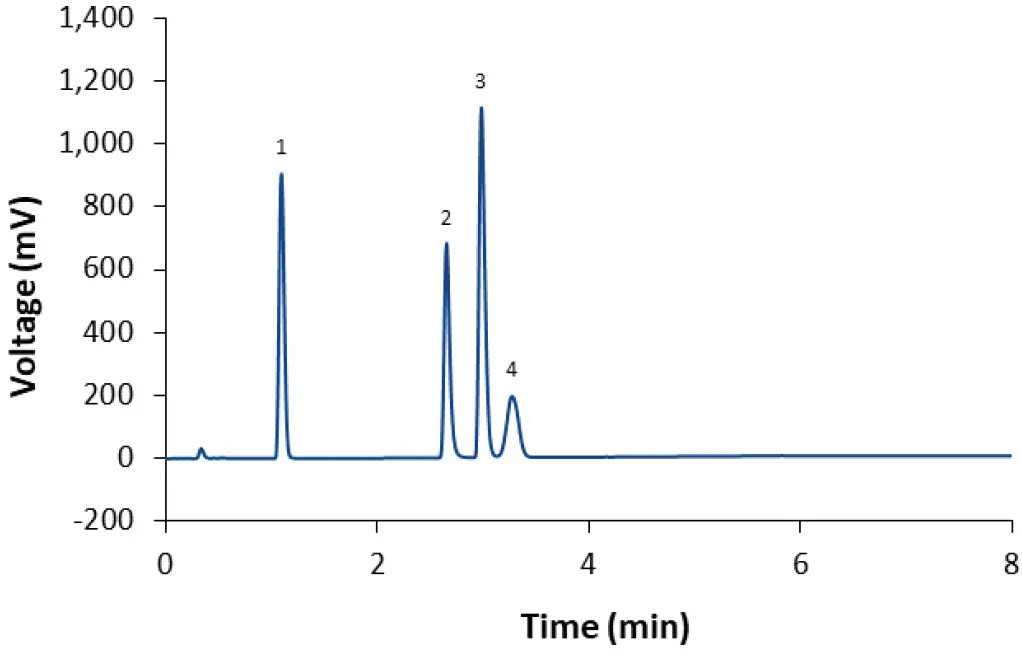
Fig. 3 Separation of LNP-0315 mix using core-shell phenyl-hexyl column and gradient M2 (90 % methanol 90 % acetonitrile in 4 min), CT = 50°C; ET = 45°C. 1: Cholesterol; 2: DSPC; 3: ALC-0315; 4: ALC-0159.
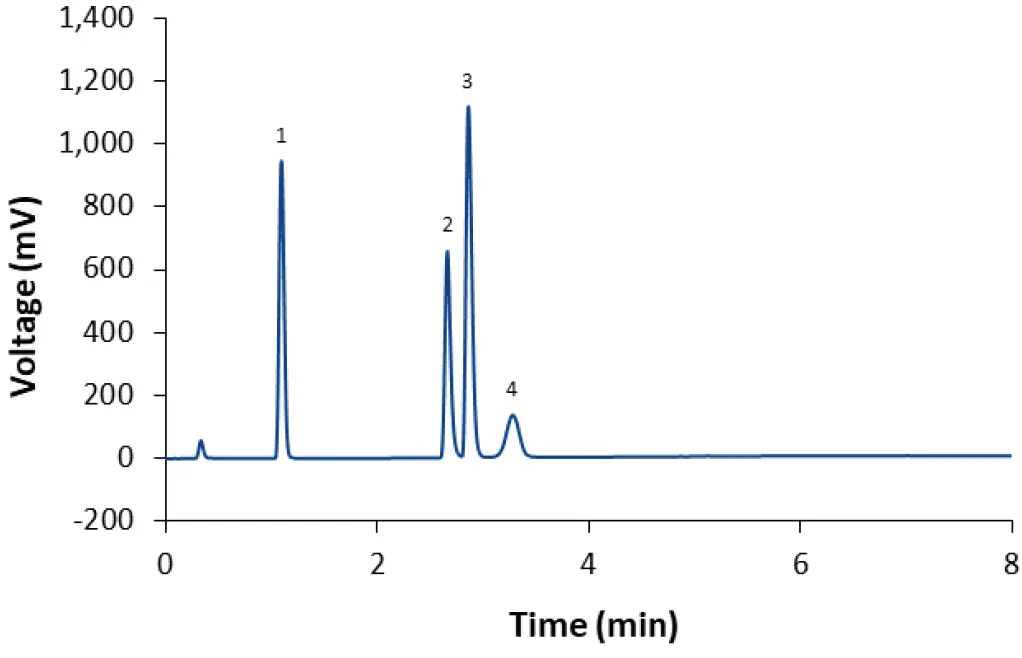
Fig. 4 Separation of LNP-SM102 mix using core-shell phenyl-hexyl column and gradient M2 (90 % methanol 90 % acetonitrile in 4 min) CT = 50°C; ET = 45°C. 1: Cholesterol; 2: DSPC; 3: SM-102; 4: DMG-PEG(2000).
Alternatively, the advantages of both solvents can be combined by using a mixture of acetonitrile and methanol as eluent. With this approach, all tested lipids can be eluted as well, and the peak shape is further improved (Fig. 5). As the acetonitrile:methanol and water:ammonium acetate ratios are constant in this quaternary gradient, the method can be transferred to a binary pump by pre-mixing the organic solvents and diluting the modifier in water.
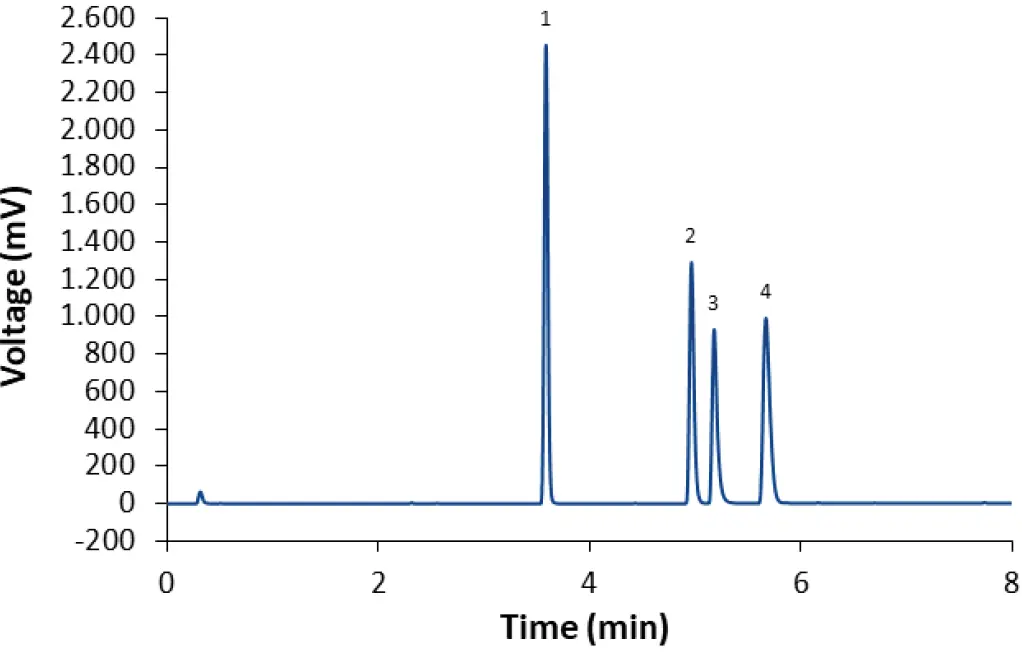
Fig. 5 Separation of LNP-0315 mix using core-shell phenyl-hexyl column and a gradient with acetonitrile:methanol 65:35 (70 % 90 % in 4 min). CT = 50°C; ET = 45°C; 0.4 ml/min; 10 mM ammonium acetate as modifier. 1: Cholesterol; 2: ALC-0159; 3: DSPC; 4: ALC-0315.
For further optimization, the modifier was investigated. For detection with an ELSD, only volatile modifiers are suitable. When no modifier was used, the ionizable lipid ALC-0315 did not elute. When formic acid was used as a modifier, all lipids were eluted, but the peak shape of ALC-0315 and DSPC was deteriorated and retention of ALC-0315 co-eluted with Cholesterol (Fig. 6). Accordingly, ammonium acetate is more suitable as a modifier for separation of the lipids. Ammonium acetate solution has a pH of 7, at which the ionizable lipid ALC-0315 is also expected to be neutral in charge [6], which appears to be advantageous for both retention and peak shape of the ionizable lipid.
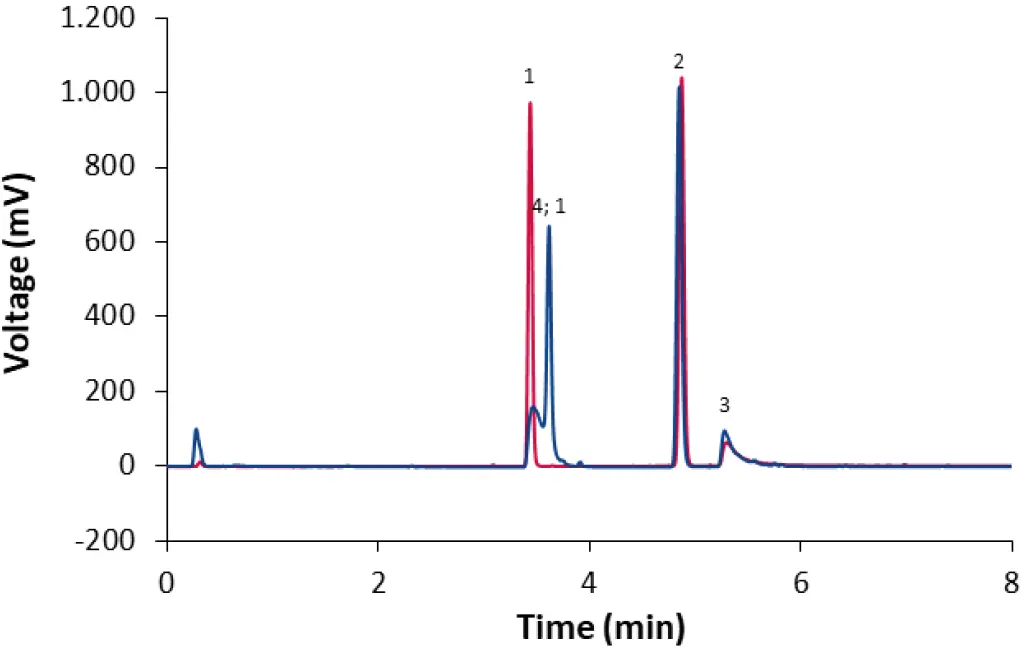
Fig. 6 Separation of LNP-0315 mix using core-shell phenyl-hexyl column and a gradient with acetonitrile:methanol 65:35 (70 % 90 % in 4 min). CT = 50°C; ET = 45°C; 0.4 ml/min; dark blue: 0.01 % formic acid as modifier; red: no modifier. 1: Cholesterol; 2: ALC-0159; 3: DSPC; 4: ALC-0315.
When the gradient method with ammonium acetate as modifier is used, three of the four lipids elute during the isocratic hold after 4 min (Fig. 5). This indicates that separation may be achieved using an isocratic method as well. After slight modifications of the eluent composition and column temperature, the lipids can indeed be separated within 3 min in an isocratic run (Fig. 7). The same method can be used to separate the lipids used in the Covid-19 vaccine developed by Moderna (Fig. 8). This approach results in earlier elution than the gradient method M2. The analysis time is greatly reduced because the column does not have to be re-equilibrated after each run. For measuring the lipid content of LNPs, the isocratic method is equally suitable as the gradient methods, while saving time and eluents.
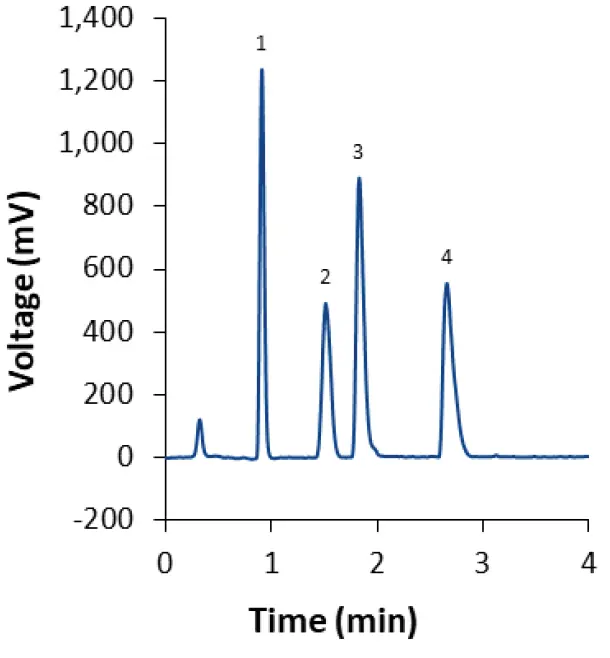
Fig. 7 Separation of LNP-0315 mix using core-shell phenyl-hexyl column and isocratic eluent composition M1. CT = 55°C; ET = 45°C; 0.4 ml/min.
1: Cholesterol; 2: ALC-0159; 3: DSPC; 4: ALC-0315.
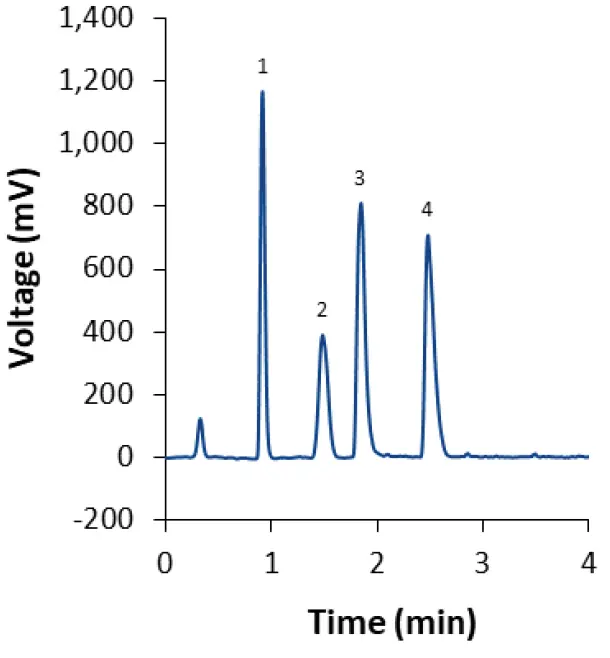
Fig. 8 Separation of LNP-SM102 mix using core-shell phenyl-hexyl column and isocratic eluent composition M1. CT = 55°C; ET = 45°C. 1: Cholesterol; 2: DMG-PEG(2000); 3: DSPC; 4: SM-102.
Optimization of ELSD temperature
The eluents of both methods for the separation of lipids from LNP formulations using the core-shell phenyl-hexyl column contain a constantly high concentration of 85 % or 90 % organic solvent, respectively. Therefore, the evaporation temperature of the ELSD can be reduced, which can result in higher sensitivity for volatile or heat-sensitive compounds. In this application, the sensitivity for Cholesterol and DSPC is indeed increased with lower evaporation temperatures. However, reducing the evaporation temperature below a threshold increases the noise because the eluent is not evaporated completely, which causes high scattering by water droplets. Under the tested conditions, this effect occurs at evaporation temperatures ≤ 33°C. The optimal S/N ratio for the isocratic method was reached at an evaporation temperature of 35°C (Fig. 9), which is applied for the calibration of all final methods in this report.
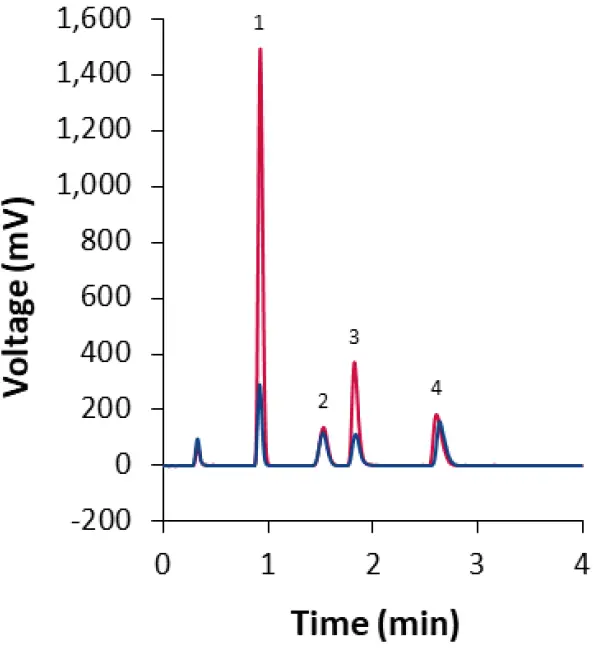
Fig. 9 Optimization of the ELSD evaporation temperature demonstrated with isocratic method M1 for the core-shell phenyl-hexyl column. Blue: ET = 45°C; Red: ET = 35°C. Sample: LNP-0315 mix (dilution 1:2). 1: Cholesterol; 2: ALC0159; 3: DSPC; 4: ALC-0315.
Calibration and Limit of Quantification (LOQ)
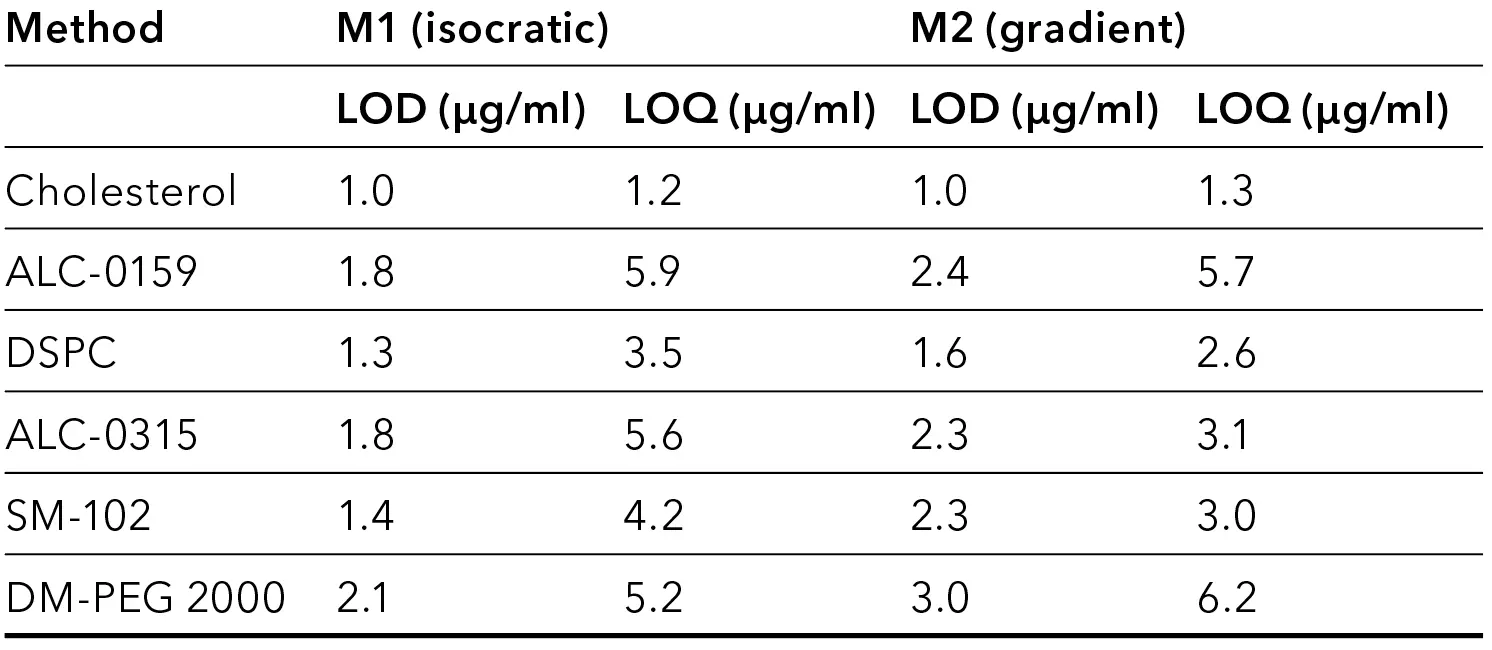
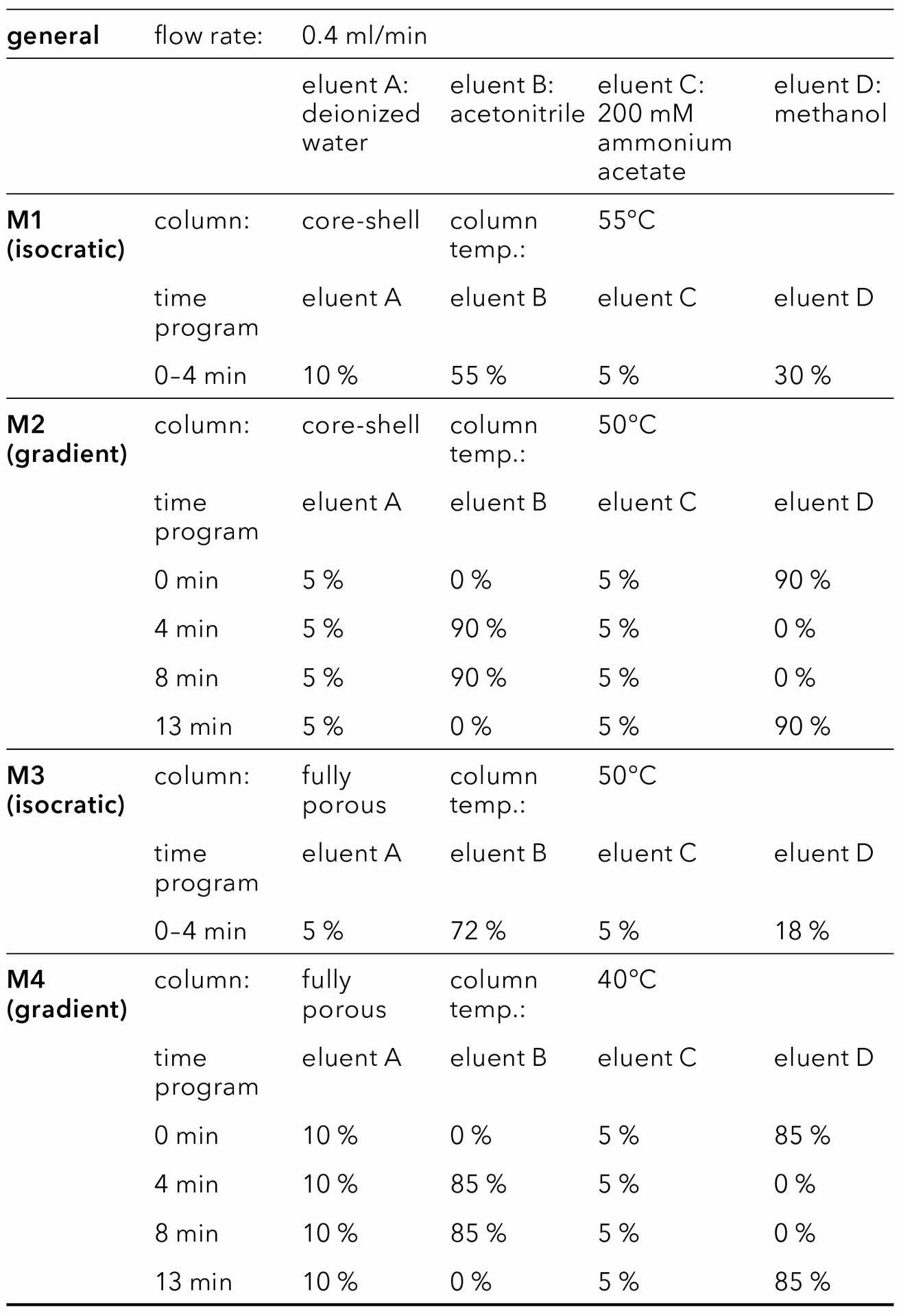
The final isocratic and gradient method were calibrated for all tested lipids. Five levels of each analyte were used for calibration. The ELSD provides linear response only for small concentration ranges. For this application, the best fit was achieved using a quadratic function. A blank was subtracted from all chromatograms of the gradient method to produce a flat baseline, which simplifies integration. A satisfactory fit with R2 > 0.999 was reached for all lipids using both methods. The limit of detection (LOD) and limit of quantification (LOQ) were approximated linearly from the peak heights at the three lowest detected levels and the ASTM noise measured in blanks. A signal-to-noise ratio (S/N) of S/N = 3 for LOD and S/N = 10 for LOQ were assumed. LODs are in the range of 1.0–3.0 µg/ml (5–15 ng), and LOQs in the range of 1.0–6.2 µg/ml (6–31 ng). LOD and LOQ of most analytes is slightly lower for the isocratic method (Tab. 2). The comparison with results from literature shows that the reached LODs and LOQs are sufficiently low. In a study on lipid stability in LNPs reported by Merck, Cholesterol and DSPC were analyzed using HPLC separation and a charged aerosol detector. With the same injection volume (5 µl), the LOQs were 36 µg/ml and 25 µg/ml, respectively, and therefore one order of magnitude higher than shown here [7].
Tab. 2 LOD and LOQ for optimized methods using the core-shell phenyl-hexyl column.
Method transfer to a fully porous column
Core-shell phases are known to improve efficiency and resolution, with less impact on system backpressure. As column backpressure is less of a concern for the small column dimension and moderate flow rates of the methods developed here, a fully porous stationary phase can be considered as well. Therefore, as an alternative, the ChromCore phenyl-hexyl UHPLC column with similar particle and pore size and the same column dimension was tested for the separation of both lipid mixtures. Initially, the gradient method optimized for the core-shell column was directly transferred. Because of the higher surface area of the fully porous column material (300 m2/g instead of 200 m2/g), the fully porous column retains all analytes stronger (Fig. 10).
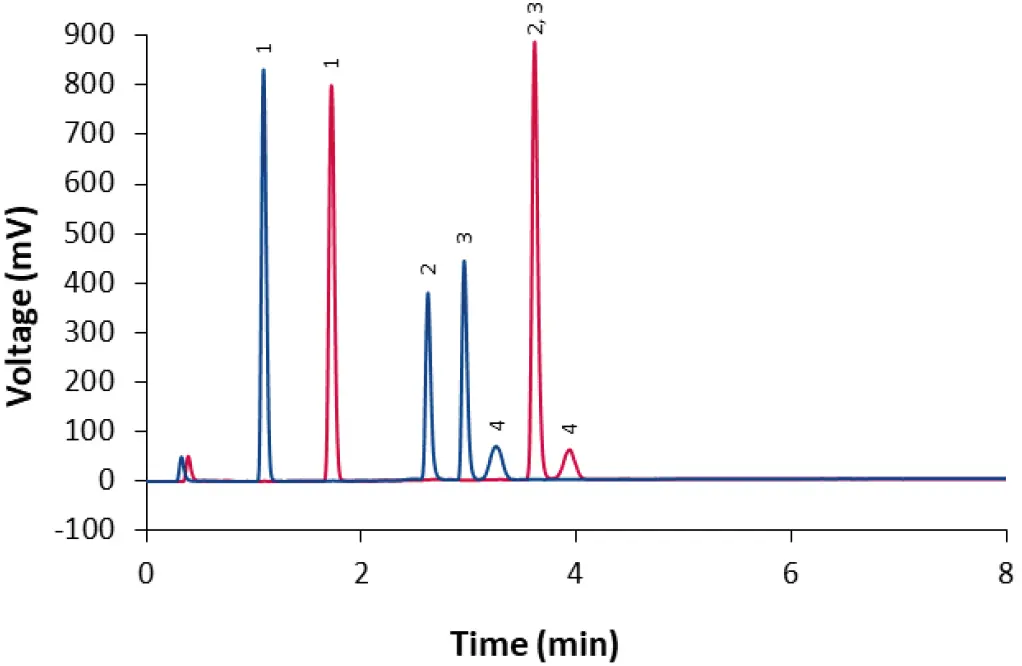
Fig. 10 Comparison of core-shell column (dark blue) and fully porous column (red) with method M2. Sample: LNP-0315 mix (dilution 1:2). 1: Cholesterol; 2: DSPC; 3: ALC-0315; 4: ALC-0159.
By increasing the water content and reducing the column temperature, lipid retention is further increased, and all analytes of each formulation can be separated (Fig. 11, Fig. 12). Notably, this also alters the peak order: DSPC is now eluted later than the PEGylated lipids.
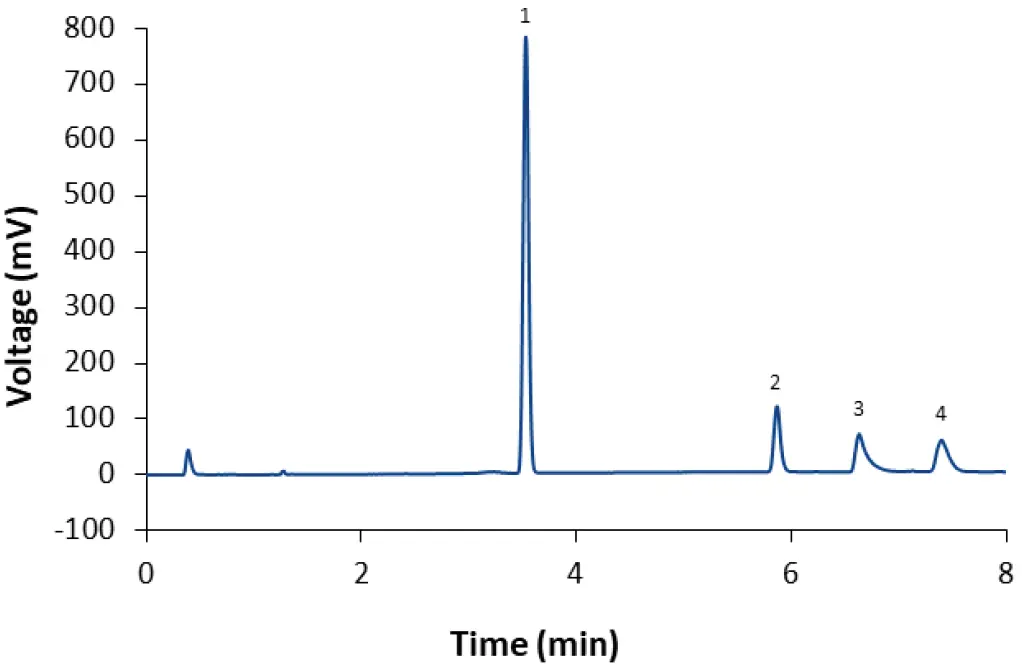
Fig. 11 Separation of LNP-0315 mix (dilution 1:2) with fully porous phenyl-hexyl column and method M4. 1: Cholesterol; 2: ALC-0159; 3: DSPC; 4: ALC-0315.
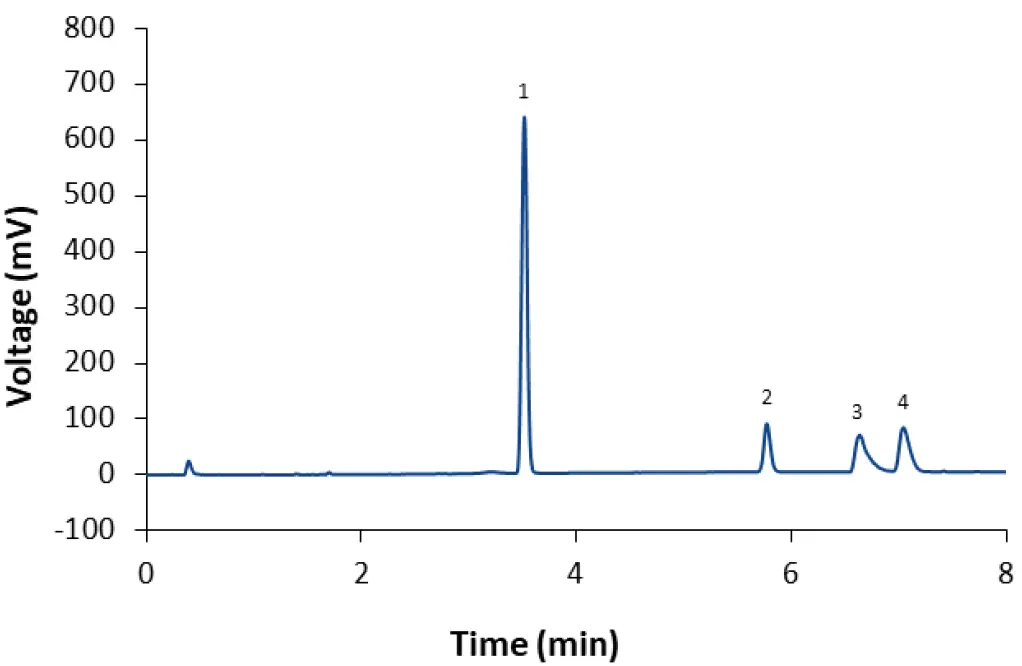
Fig. 12 Separation of LNP-SM102 mix (dilution 1:2) with fully porous phenyl-hexyl column and method M4. 1: Cholesterol; 2: DMG-PEG(2000); 3: DSPC; 4: SM-102.
After slight modification of the isocratic method, the baseline separation of the lipids with the fully porous column was achieved in around 2 min (Fig. 13, Fig. 14). Calibration of both methods optimized for the fully porous column results in good fits with R2 ≥ 0.999 for all analytes. LOD and LOQ values were further improved in comparison to the core shell column (Tab. 3).
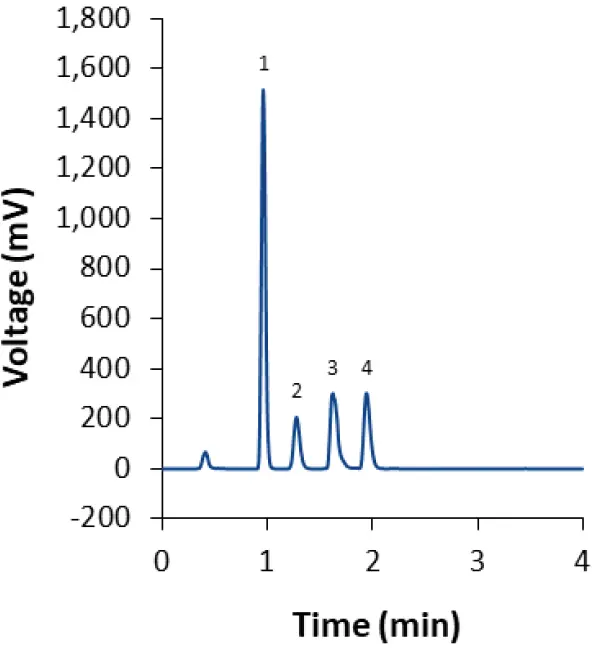
Fig. 13 Separation of of LNP-0315 mix (dilution 1:2) with fully porous phenyl-hexyl column and isocratic method M3. 1: Cholesterol; 2: ALC-0159; 3: DSPC; 4: ALC-0315.
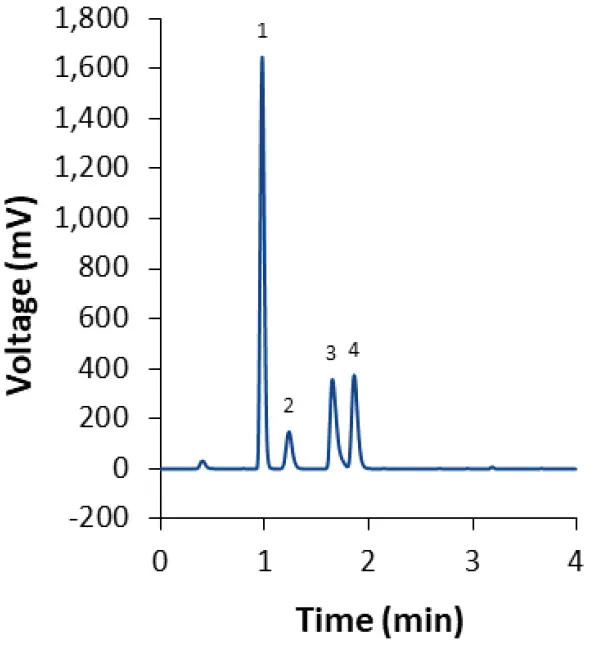
Fig. 14 Separation of lipids LNP-SM102 mix (dilution 1:2) with fully porous phenyl-hexyl column isocratic method M3. 1: Cholesterol; 2: DM-PEG 2000; 3: DSPC; 4: SM-102.
Tab. 3 LOD and LOQ for optimized methods using the fully porous phenyl-hexyl column.
Conclusion
Two phenyl-hexyl columns were successfully used for the separation of two different lipid mixtures, which are relevant for LNP formulations. The need to combine acetonitrile and methanol as eluents and to add ammonium acetate as a modifier for good peak shape and complete elution was demonstrated. Two method options were established for both columns. The shown gradient methods are slower, but offer more space to separate possible impurities as well. The isocratic methods are equally suitable for assaying the lipid content of LNP formulations and are much faster. For each analyte, LOD and LOQ were similar for all methods. The ELSD provides high sensitivity for all lipids with LODs in the range of 0.8–3 µg/ml or 4–15 ng. LOQs of 0.9–4 µg/ml or 5–32 ng were reached.
Material and Methods
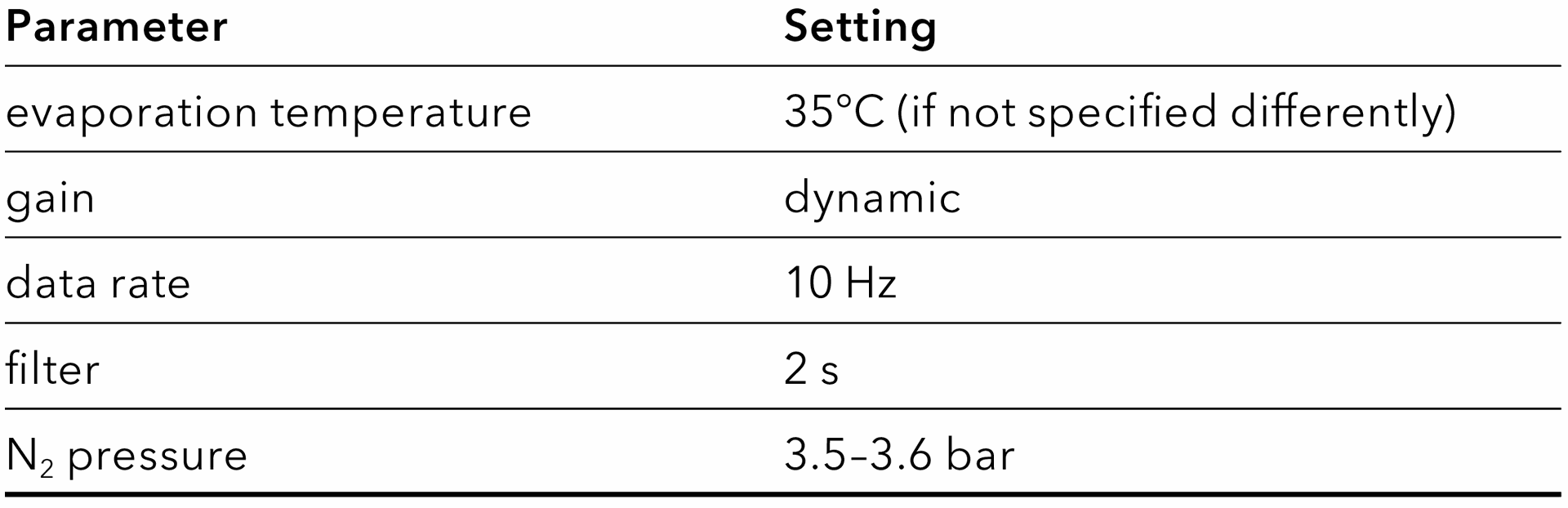
Tab. 4 Pump and column parameters of final methods.
Tab. 5 Settings of the evaporative light scattering detector.
Tab. 6 Instruments
|
Instrument |
Description |
Article No. |
|
pump |
AZURA P 6.1L, LPG |
|
|
autosampler |
AZURA AS 6.1L |
|
|
detector |
SEDEX 100LT |
|
|
thermostat |
AZURA CT 2.1 |
|
|
core-shell column |
Kinetex® 1.7 µm phenyl-hexyl 100 Å; 2.1x50 mm (Phenomenex Part No 00B-4500-AN) |
– |
|
fully porous column |
ChromCore 1.8 µm phenyl-hexyl 120 Å; 2.1x50 mm (Nanochrom Part No A311-018012-02105S) |
– |
|
software |
ClarityChrom 9.0.0 |
References
[1] Hald Albertsen, C., Kulkarni, J. A., Witzigmann, D., Lind, M., Petersson, K., Simonsen, J. B. Adv Drug Deliv Rev 2022, 188, 114416.
[2] https://www.usp.org/mrna: Analytical Procedures for mRNA Vaccine Quality. Draft guidelines: 2nd Edition. reviewed 16.01.2024
[3] 5994-6051EN. Schneider, S., ©Agilent Technologies, Inc. 2023
[4] 720007331. Alden, B. A., Isaac, G., Chen, W., Lauber, M. A.; ©Waters Corporation. 2021
[5] 5994-4709EN. Schneider, S., ©Agilent Technologies, Inc. 2022
[6] Higuchi, A., Sung, T. C., Wang, T., Ling, Q. D., Kumar, S. S., Hsu, S. T., Umezawa, A. Polymer Reviews 2023, 63, 394–436.
[7] Kinsey, C., Lu, T., Deiss, A., Vuolo, K., Klein, L., Rustandi, R. R., Loughney, J. W. Electrophoresis 2022, 43, 1091–1100.
Application details
Method | HPLC |
Mode | RP |
Substances | Cholesterol, ALC-0159, DSPC, ALC-0315, SM-102, DM-PEG 2000 |
CAS number | 57-88-5, 816-94-4, 2036272-55-4, 1849616-42-7, 2089251-47-6, 160743-62-4 |
Version | Application No.: VPH0078 Version 1 / 03/2024 | ©KNAUER Wissenschaftliche Geräte GmbH |

